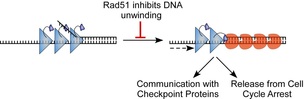
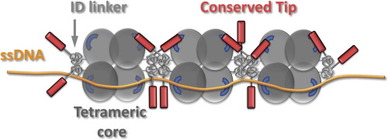
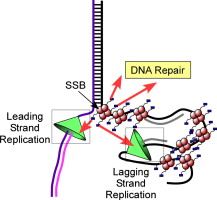
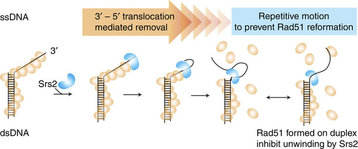
|
50. Kashyap R., Deveryshetty D., Walsh N., Tokmina-Lukaszewska M., Bothner B., Bennett B., Antony E. Cryo-EM captures the coordination of long-range allostery and asymmetric electron transfer by a bi-copper cluster in the nitrogenase-like DPOR complex
bioRxiv 2024.04.26.590571; doi: https://doi.org/10.1101/2024.04.26.590571 |
|
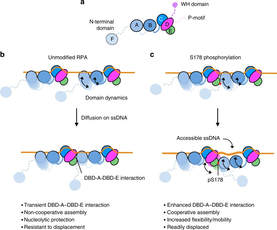
49. Chadda R., Kaushik V., Ahmad I.M., Deveryshetty J., Holehouse A., Sigurdsson S.T., Bothner B., Dastvan R., Origanti S., and Antony E. Wrapping of single-stranded DNA by Replication Protein A and modulation through phosphorylation. bioRxiv 2024.03.28.587234; doi: https://doi.org/10.1101/2024.03.28.587234
|
48. Granger L.S., Sharma S., Kaushik V., Razzaghi R., Honda M., Bhat D.S., Wlodarski M., Antony E., and Spies M. Human hnRNPA1 reorganizes telomere-bound Replication Protein A.
bioRxiv 2023.05.09.540056; doi: https://doi.org/10.1101/2023.05.09.540056
bioRxiv 2023.05.09.540056; doi: https://doi.org/10.1101/2023.05.09.540056
47. Norris J.L., Rogers L.O., Pytko K.G., Dannenberg R.L., Perreault S., Kaushik V., Kuppa S., Antony E., and Hedglin M. Interplay of macromolecular interactions during assembly of human DNA polymerase δ holoenzymes and initiation of DNA synthesis. bioRxiv 2023.05.09.539896; doi: https://doi.org/10.1101/2023.05.09.539896. In Press. Nucleic Acids Research.
|
46. Pangeni S., Biswas G., Kaushik V., Kuppa S., Yang O., Chang-Tin L., Mishra G., Levy Y., Antony E., and Ha T.J. Rapid long-distance migration of RPA on single stranded DNA occurs through intersegmental transfer utilizing multivalent interactions.
J. Mol. Biol. 2024. doi: 10.1016/j.jmb.2024.168491 |
|
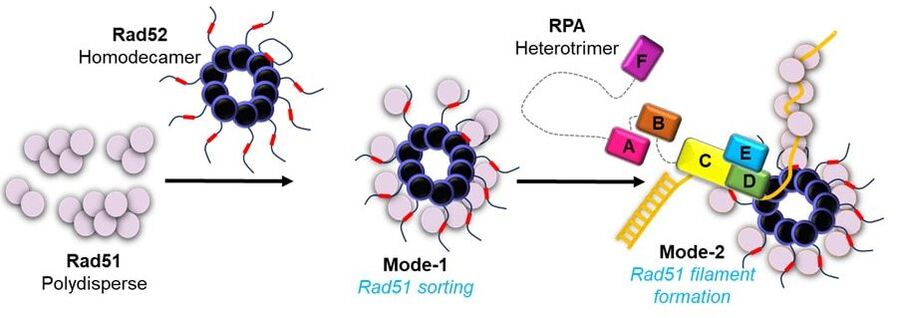
43. Deveryshetty J., Chadda R., Mattice J., Karunakaran S., Rau M.J., Basore K., Pokhrel N., Englander N., Fitzpatrick J, A., Bothner B., and Antony E. C-terminus induced asymmetry within a Rad52 homodecamer dictates single-position Rad51 nucleation in homologous recombination.
Nature Communications. 2023. 14(1):6215. doi: 10.1038/s41467-023-41993-1. |
|
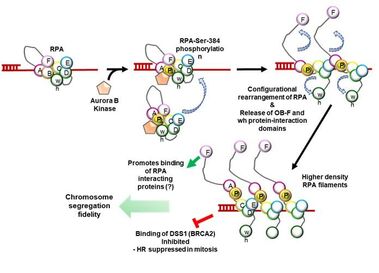
42. Roshan P., Kuppa S., Mattice J., Kaushik V., Chadda R., Pokhrel N., Tumala B., Biswas A., Bothner B., Antony E., and Origanti S. An Aurora B-RPA signaling axis secures chromosome segregation fidelity.
Nature Communications. 2023. 14(1):3008. doi: 10.1038/s41467-023-38711-2. |
|
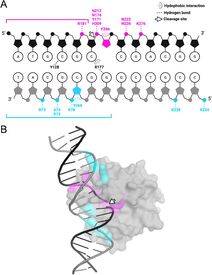
41. Hoitsma N.M., Norris J., Khoang T.H., Kaushik V., Antony E., Hedglin M., and Freudenthal B.D. Mechanistic insight into AP-Endonuclease I cleavage of abasic sites at stalled replication forks.
Nucleic Acids Research. 2023. 51(13):6738. DOI: 10.1093/nar/gkad481 |
|
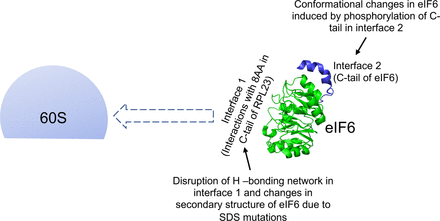
40. Eliff J., Biswas A., Kuppa S., Roshan P., Patterson A., Mattice J., Chinnaraj M., Burd R., Walker S.E., Pozzi N., Antony E., Bothner B., and Origanti S. Dynamic states of eIF6 and SDS variants modulate interaction with uL14 of the 60S robosomal subunit.
Nucleic Acids Res. 2023. 51(4):1803-1822.doi: 10.1093/nar/gkac1266. |
|
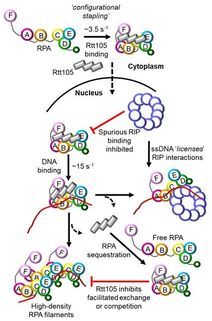
39. Kuppa S., Deveryshetty J., Chadda R., Mattice J., Pokhrel H., Kaushik V., Patterson A., Dhingra N., Pangeni S., Sadauskas M.K., Shiekh S., Balci H., Ha T., Zhao X., Bothner B., and Antony E. Rtt105 configurationally staples RPA and blocks facilitated exchange and interactions with RPA-interacting proteins.
Nature Communications. 2022. 13(1):5152. doi: 10.1038/s41467-022-32860-6. |
37. Hormeno S, Wilkinson O.J., Aicart-Ramos C., Kuppa S., Antony E. , Dillingham M.S., and Moreno-Herrero F. Human HELB is a processive motor protein which catalyses RPA clearance from single-stranded DNA.
Proc Natl Acad Sci U S A. 2022 12;119(15):e2112376119. doi: 10.1073/pnas.2112376119
Proc Natl Acad Sci U S A. 2022 12;119(15):e2112376119. doi: 10.1073/pnas.2112376119
|
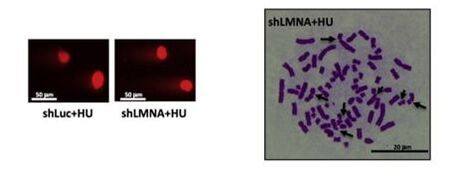
38. Graziano S., Coll-Bonfill N., Teodoro-Castro B., Kuppa S., Jackson J., Shaskova E., Mahajan U., Vindigni A., Antony E., and Gonzalo S. A-type lamins are critical for the recruitment of RPA and RAD51 to stalled replication forks to maintain fork stability.
J. Biological Chemistry. 2021. 297(5):101301.www.sciencedirect.com/science/article/pii/S0021925821011078 |
|
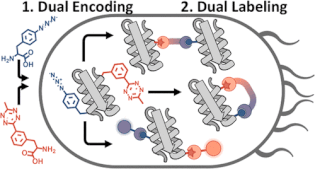
36. Bednar R.M., Jana S., Kuppa S., Franklin R., Beckman J., Antony E., Cooley R.B., Mehl R.A.. Genetic Incorporation of Two Mutually Orthogonal Bioorthogonal Amino Acids That Enable Efficient Protein Dual-Labeling in Cells.
ACS Chemical Biology. 2021 https://pubs.acs.org/doi/10.1021/acschembio.1c00649. |
|
34. Kuppa S., Pokhrel N., Corless E., Origanti S., and Antony E. Generation of fluorescent versions of Saccharomyces cerevisiae RPA to study the conformational dynamics of its ssDNA-binding domains.
Methods Mol. Biol. 2021. doi: 10.1007/978-1-0716-1290-3_9. https://link.springer.com/protocol/10.1007%2F978-1-0716-1290-3_9 |
|
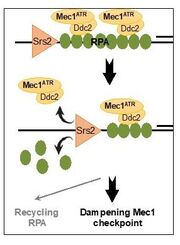
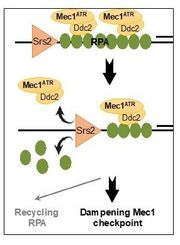
33. Dhingra, N., Kuppa, S., Wei, L., Pokhrel, N., Baburyan, S., Meng, X., Antony, E., and Zhao, X. The Srs2 helicase dampens DNA damage checkpoint by recycling RPA from chromatin. Proc Natl Acad Sci U S A. 2021. 119(15):e2112376119. doi: 10.1073/pnas.2020185118
https://www.pnas.org/content/118/8/e2020185118 |
|
32. Ahmad, F., Patterson, A., Deveryshetty, J., Mattice, J., Pokhrel, N., Bothner, B., and Antony, E. Hydrogen-deuterium exchange reveals a dynamic DNA binding map of Replication Protein A.
Nucleic Acids Research. 2021. 49(3):1455-1469. doi: 10.1093/nar/gkaa1288. https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkaa1288/6097544 |
|
31. Corless, E.I., Imran S.M.S., Watkins, M.B., Bacik, J.P., Mattice J., Patterson A., Danyal, L.C., Soffe, M., Kitelinger, R., Seefeldt L.C., Origanti, S., Bennett, B., Ando, A., and Antony, E. The flexible N-terminus of BchL autoinhibits activity through interaction with its [4Fe-4S] and relieved upon ATP binding.
J. Biol. Chem. 2020. doi: 10.1074/jbc.RA120.016278. LINK. |
30. Quinn, S.M., Vargason, T., Pokhrel, N., Antony, E., Hahn, J., and Gilbert S.P. KIF3A accelerates KIF3C within the kinesin-2 heterodimer to generate symmetrical phosphate release rates for each processive step.
J. Biol. Chem. 2020. doi: 10.1074/jbc.RA120.015272. LINK.
J. Biol. Chem. 2020. doi: 10.1074/jbc.RA120.015272. LINK.
|
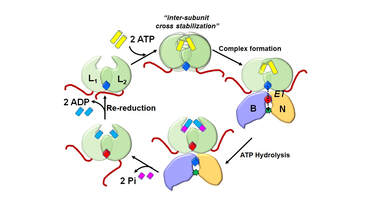
29. Corless, E.I., Bennet, B.B., and Antony, E. Substrate recognition induces sequential electron transfer across subunits in the nitrogenase-like DPOR complex. Journal of Biological Chemistry.
J Biol Chem. 2020. 295(39):13630-13639. doi: 10.1074/jbc.RA120.015151. LINK. |
28. Corless, E. I., Mettert, E. L., Kiley, P. J. and Antony, E. Elevated Expression of a Functional Suf Pathway in the E.coli BL21(DE3) Cell Line Enhances Recombinant Production of an Iron-Sulfur Cluster Containing Protein.
J Bacteriol. 2020. 202(3):e00496-19. doi: 10.1128/JB.00496-19. LINK.
J Bacteriol. 2020. 202(3):e00496-19. doi: 10.1128/JB.00496-19. LINK.
|
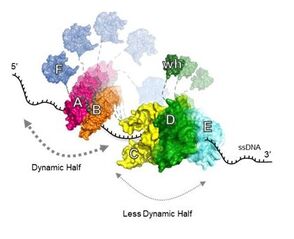
27. Pokhrel N., Caldwell, C.C., Corless, E.I., Tillison, E.A., Tibbs, J., Jocic, N., Ali Tabei, S.M., Wold, M.S., Spies, M. and Antony E. Dynamics and Selective Remodeling of the DNA Binding Domains of RPA.
Nature Structural & Molecular Biology. 2019. 26, p129–136 (2019). https://www.nature.com/articles/s41594-018-0181-y Featured as cover |
|
26. Yates LA, Aramayo RJ, Pokhrel N, Caldwell CC, Kaplan JA, Perera RL, Spies M, Antony E, and Zhang X. A structural and dynamic model for the assembly of Replication Protein A on single stranded DNA.
Nature Communications. 2018. doi: 10.1038/s41467-018-07883-7 |
|
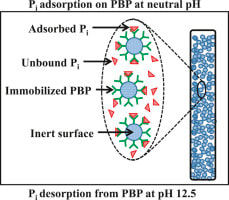
25. Venkiteshwaran K., Pokhrel N., Hussein F., Antony E., and Mayer B.K. Phosphate removal and recovery using phosphate binding proteins.
Water Research X. 2018. https://doi.org/10.1016/j.wroa.2018.09.003 |
24. Mustafa G., Chuang C. , Roy W.A., Farhath M.A., Pokhrel N., Ma Y., Nagasawa K., Antony E., Comstock M.J., Basu S., and Balci H. A Force Sensor that Converts Fluorescence Signal into Force Measurement Utilizing Short Looped DNA.
Biosensors and Bioelectronics. 2018. doi: 10.1016/j.bios.2018.08.073
Biosensors and Bioelectronics. 2018. doi: 10.1016/j.bios.2018.08.073
|
23. Seefeldt L., Hoffman B., Peters J., Raugei S., Antony E., and Dean D. Energy Transduction in Nitrogenase.
Accounts of Chemical Research. 2018. doi: 10.1021/acs.accounts.8b00112 |
|
22. Singh S.P., Kukshal V., De Bona P., Antony E., and Galletto R. The mitochondrial single-stranded DNA binding protein from S. cerevisiae, Rim1, does not form stable homo-tetramers and binds DNA as a dimer of dimers. Nucleic Acids Research. 2018. doi: 10.1093/nar/gky530
|
|
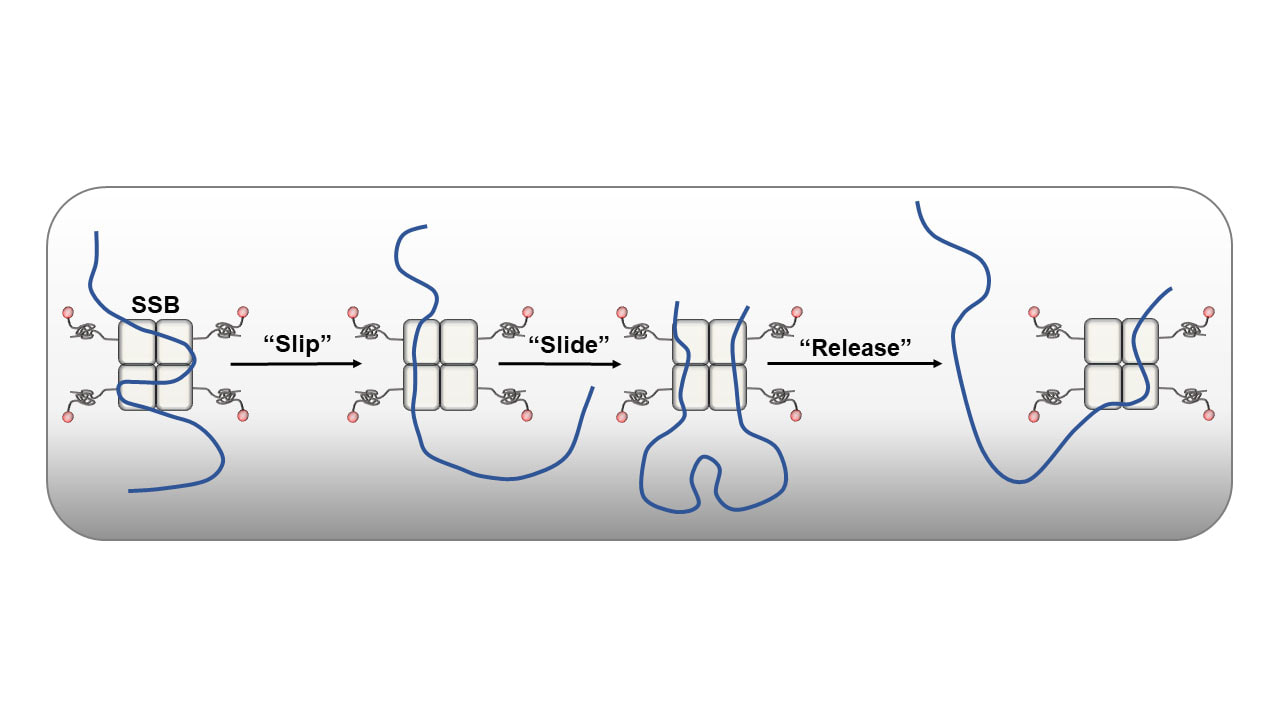
21. Antony E., and Lohman T.M. Dynamics of E. coli single stranded DNA binding (SSB) protein-DNA complexes.
Seminars in Cell and Developmental Biology. 2018. S1084-9521(17)30432-9. DOI: 10.1016/j.semcdb.2018.03.017 |
|
20. Pokhrel N., Origanti S., Davenport E.P., Gandhi D., Kaniecki K., Mehl R., Greene E., Dockdendorff C., and Antony E. Monitoring Replication Protein A (RPA) dynamics through site-specific incorporation of non-canonical amino acids.
Nucleic Acids Research. 2017. 45(16):9413-9426. DOI: https://doi.org/10.1093/nar/gkx598 |
|
19. Danyal, K., Shaw, S., Page, T.P., Duval, S., Fielding, A.J., Horitani, M., Marts, A.R., Lukoyanov, L., Dean, D.R., Raugei, S., Hoffman, B.M., Seefeldt, L.C. and Antony E. Negative Cooperativity in the Nitrogenase Fe Protein Electron Delivery Cycle.
PNAS. 2016. doi: 10.1073/pnas.1613089113 |
|
18. Davenport E.P., Harris D.F., Origanti S., and Antony E. Rad51 nucleoprotein filament disassembly captured using fluorescent Plasmodium falciparum SSB as a reporter for single-stranded DNA. PLos One. 2016.
http://dx.doi.org/10.1371/journal.pone.0159242 |
|
17. Yang Z.Y., Lebbetter R., Shaw S., Pence N., Tokmina-Lukaszewska M., Eilers B., Guo Q., Pokhrel N., Cash V.L., Dean D.R., Antony E., Bothner B., Peters J.W., and Seefeldt L.C. Evidence that the Pi release event is the rate limiting step in the nitrogenase catalytic cycle. Biochemistry. 2016. 5:55(26):3625-3635.
|
16. Burgess J., Jones H.B., Kumar P., Toth R., Middaugh C.R., Antony E.* and Dickenson N.E.*. Spa47 is an oligomerization-activated type three secretion system (T3SS) ATPase from Shigella flexneri. Protein Science. 2016. 25(5):1037-48. (*Co-Corresponding Authors)
|
15. Duval S., Danyal K., Shaw S., Dean D.R., Hoffman B.M., Antony E*. and Seefeldt L.C.* Establishing the order of electron transfer and ATP hydrolysis in Nitrogenase . PNAS. 2013. 110:16414-16419 (*Co-Corresponding Authors).
Highlighted in Science USU Press Release |
Postdoctoral and Graduate Publications
|
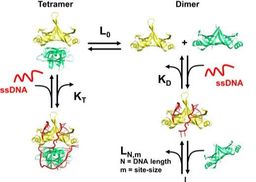
10. Antony E., Kozlov A.J., Nguyen B., and Lohman T.M. Plasmodium falciparum SSB tetramer binds single-stranded DNA only in a fully wrapped mode. J. Mol. Biol. 2012. 420: 284-295. Featured as cover 9. Antony E., Weiland E.A., Korolev S., and Lohman T.M. Plasmodium falciparum SSB tetramer wraps single-stranded DNA with similar topology but opposite polarity to E. coli SSB. J. Mol. Biol. 2012. 420: 269-283. |
7. Antony E., and Lohman T.M. "Non-hexameric SF1 DNA helicases and translocases". Encyclopedia of Biological Chemistry, (2nd edition) W.J. Lennarz and M.D. Lane, eds. Elsevier Science (2010).
5. Antony E., Khubchandani S., Chen S., and Hingorani M.M. Contribution of Msh2 and Msh6 subunits to the asymmetric ATPase and DNA mismatch binding activities of Saccharomyces cerevisiae Msh2-Msh6 mismatch repair protein. DNA Repair. 2006. 5(2):153-62.
4. Zito C.R. *, Antony E.*, Hunt J.F., Oliver D.B., and Hingorani M.M. Role of a conserved glutamate residue in the Escherichia coli SecA ATPase mechanism. J. Biol. Chem. 2005. 280:14611-14619 (* Co-first authors).
3. Antony E. and Hingorani M.M. Asymmetric ATP binding and hydrolysis activity of the Thermus aquaticus MutS dimer is key to modulation of its interactions with mismatched DNA. Biochemistry. 2004. 43:13115-13128.
2. Antony E. and Hingorani M.M. Mismatch recognition-coupled stabilization of Msh2-Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry. 2003. 42:7682-93.
1. Finkelstein J., Antony E., Hingorani M.M., and O'Donnell M. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Anal. Biochem. 2003. 319:78-87.
4. Zito C.R. *, Antony E.*, Hunt J.F., Oliver D.B., and Hingorani M.M. Role of a conserved glutamate residue in the Escherichia coli SecA ATPase mechanism. J. Biol. Chem. 2005. 280:14611-14619 (* Co-first authors).
3. Antony E. and Hingorani M.M. Asymmetric ATP binding and hydrolysis activity of the Thermus aquaticus MutS dimer is key to modulation of its interactions with mismatched DNA. Biochemistry. 2004. 43:13115-13128.
2. Antony E. and Hingorani M.M. Mismatch recognition-coupled stabilization of Msh2-Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry. 2003. 42:7682-93.
1. Finkelstein J., Antony E., Hingorani M.M., and O'Donnell M. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Anal. Biochem. 2003. 319:78-87.